Class 12 Biology Chapter 6 Molecular Basis of Inheritance Notes: Access comprehensive notes and solutions for Class 12 Biology Chapter 6 on Molecular Basis of Inheritance. Dive into the intricate world of DNA, RNA, gene expression, and genetic engineering with detailed explanations, diagrams, and key concepts to deepen your understanding of molecular genetics.
Class 12 Biology Chapter 6 Molecular Basis of Inheritance Notes
The DNA: DNA (Deoxyribonucleic Acid) and RNA (Ribonucleic Acid) are two types of nucleic acid found in living organisms. DNA acts as genetic material in most of the organisms. RNA also acts as genetic material in some organisms as in some viruses and acts as messenger. It functions as adapter, structural, and in some cases as a catalytic molecule.
The DNA it is a long polymer of deoxyribonucleotides. A pair of nucleotides is also known as base pairs. Length of DNA is usually defined as number of nucleotides present in it. Escherichia coli have 4.6 x 106 bp and haploid content of human DNA is 3.3 × 109 bp.
Structure of Polynucleotide Chain:

A nucleotide has three components – a nitrogenous base, a pentose sugar (ribose in case of RNA, and deoxyribose for DNA), and a phosphate group. There are two types of nitrogenous bases – Purines (Adenine and Guanine), and Pyrimidines (Cytosine, Uracil and Thymine). Cytosine is common for both DNA and RNA and Thymine is present in DNA. Uracil is present in RNA at the place of Thymine.

A polynucleotide chain:
A nitrogenous base is linked to pentose sugar with N-glycosidic linkage to form to form a nucleoside. When phosphate group is linked 5’-OH of a nucleoside through phosphoester linkage nucleotide is formed. Two nucleotides are linked through 3’-5’ phosphodiester linkage to form dinucleotide. More nucleotide joins together to form polynucleotide. In RNA, nucleotide residue has additional –OH group present at 2’-position in ribose and uracil is found at the place of Thymine.
DNA & RNA
Structure differences:
| DNA | RNA |
| The sugar present in DNA is 2-deoxy-D – (-) -ribose. | The sugar present in RNAis D- (-)- ribose. |
| DNA contains cytosine and thymine as pyrimidine bases and guanine and adenine is purine bases. | RNA contains cytosine and uracil pyrimidine bones and guanine and adenine as purine bases. |
| DNA has double strand α-helix structure. | RNA has a single stranded α-helix structure. |
| DNA molecules are very large their molecular mass may vary from 6 × 106 – 16 × 106 ʯ | RNA molecules are comparatively much smaller with molecular mass ranging from 20,000 – 40,000. |
Functional differences:
DNA: DNA has unique property of replication. DNA controls the transmission of hereditary effects.
RNA: RNA usually does not replicate. RNA controls the synthesis of proteins.
DNA double helix:
In 1953 that James Watson and Francis Crick, based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed a very simple but famous Double Helix model for the structure of DNA, the ratios between Adenine and Thymine and Guanine and Cytosine are constant and equals one The base pairing confers a very unique property to the polynucleotide chains. They are said to be complementary to each other, each strand from a DNA act as a template for synthesis of a new strand, the two double stranded DNA produced would be identical to the parental DNA molecule.

The silent features of this model are:
- DNA is made of two polynucleotide chains in which backbone is made up of sugar-phosphate and bases projected inside it.
- Two chains have anti-parallel polarity. One 5’ → 3’ and with 3’ → 5’.
- The bases in two strands are paired through H-bonds. Adenine and Thymine forms double hydrogen bond and Guanine and Cytosine forms triple hydrogen bonds.
- Two chains are coiled in right handed fashion. The pitch of helix is 3.4 nm and roughly 10 bp in each turn.
- The plane of one base pair stacks over the other in double helix to confer stability.

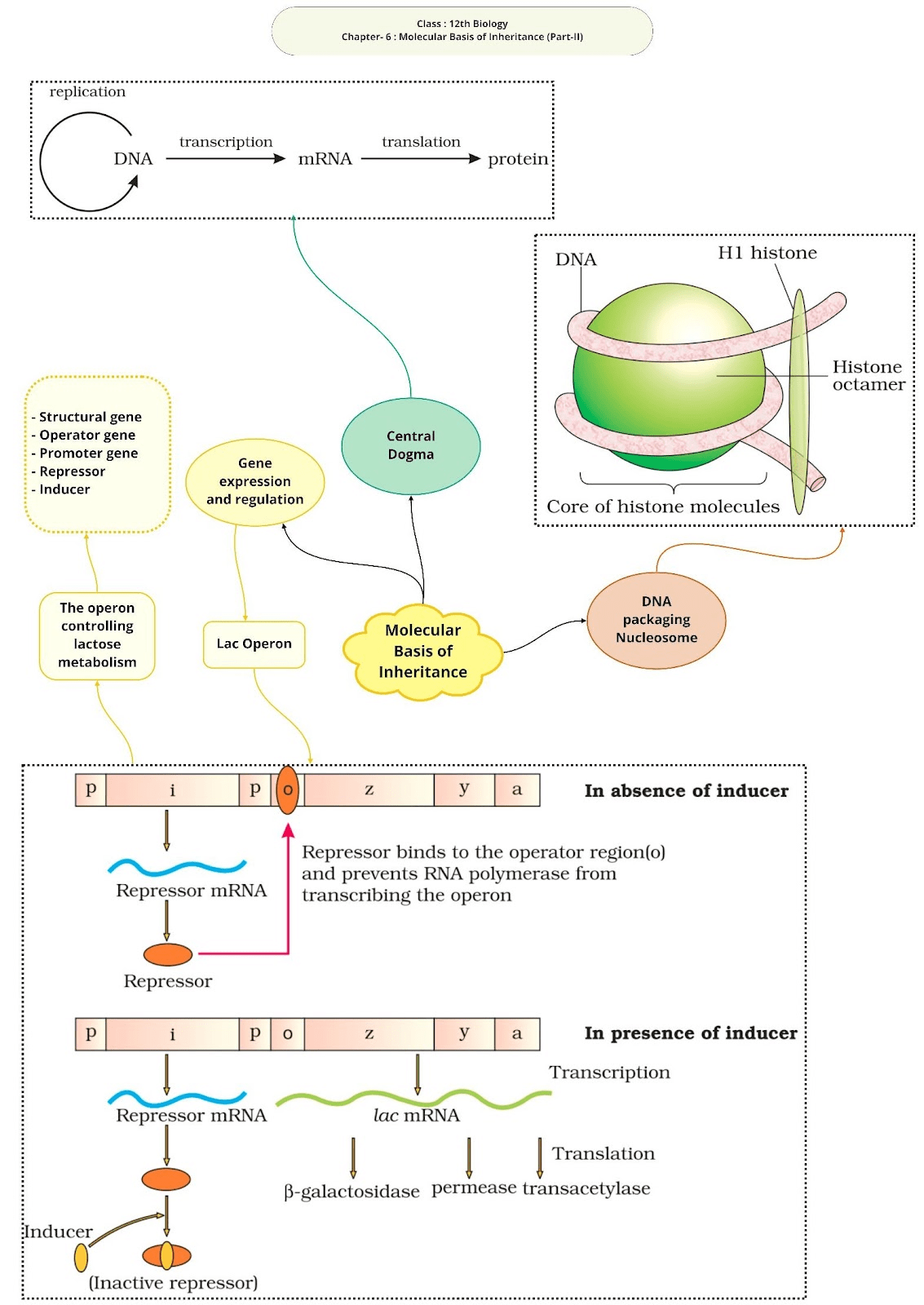
Francis Crick proposed the Central dogma in molecular biology, which states that the genetic information flows from DNA → RNA → Protein.

Packing of DNA helix:
In prokaryotes, well defined nucleus is absent and negatively charged DNA is combined with some positively charged proteins called nucleoids.
In eukaryotes, histones, positively charged protein organized to form 8 molecules unit called histone octamer. Negatively charged DNA is wrapped around the histone octamer to form nucleosome. Histones are rich in the basic amino acid residues lysine’s and arginine’s. Both the amino acid residues carry positive charges in their side chains. Single nucleosome contains about 200 base pairs. Chromatin is the repeating unit of nucleosome.
In nucleus, some region of chromatin are loosely packed (and stains light) and are referred to as euchromatin. The chromatin that is more densely packed and stains dark are called as Heterochromatin. Euchromatin is transcriptionally active chromatin, whereas heterochromatin is inactive.

Genetic Material DNA:
DNA acts as a genetic material took long to be discovered and proven. Gregor Mendel, Walter Sutton, Thomas Hunt Morgan and numerous other scientists had narrowed the search to the chromosomes located in the nucleus of most cells. In 1928, Frederick Griffith, in a series of experiments with Streptococcus pneumoniae (bacterium responsible for pneumonia), witnessed a miraculous transformation in the bacteria. When Streptococcus pneumoniae (pneumococcus) bacteria are grown on a culture plate, some produce smooth shiny colonies (S) while others produce rough colonies (R). This is because the S strain bacteria have a mucous (polysaccharide) coat, while R strain does not. Mice infected with the S strain (virulent) die from pneumonia infection but mice infected with the R strain do not develop pneumonia.

Griffith was able to kill bacteria by heating them. He observed that heat-killed S strain bacteria injected into mice did not kill them. When he injected a mixture of heat-killed S and live R bacteria, the mice died. Moreover, he recovered living S bacteria from the dead mice.

He concluded that the R strain bacteria had somehow been transformed by the heat-killed S strain bacteria. transferred from the heat-killed S strain, had enabled the R strain to synthesize a smooth polysaccharide coat and become virulent.
The Hershey-Chase experiment:
Alfred Hershey and Martha Chase (1952). They worked with viruses that infect bacteria called bacteriophages. The bacteriophage attaches to the bacteria and its genetic material then enters the bacterial cell. it was protein or DNA from the viruses that entered the bacteria. They grew some viruses on a medium that contained radioactive phosphorus and some others on medium that contained radioactive sulfur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. and viruses grown on radioactive sulfur contained radioactive protein but not radioactive DNA because DNA does not contain sulfur Radioactive phages were allowed to attach to E. coli bacteria. the viral coats were removed from the bacteria by agitating them in a blender. The virus particles were separated from the bacteria by spinning them in a centrifuge. Bacteria which was infected with viruses that had radioactive DNA indicating that DNA was the material that passed from the virus to the bacteria. Bacteria that were infected with viruses that had radioactive proteins were not radioactive. This indicates that proteins did not enter the bacteria from the viruses. DNA is therefore the genetic material that is passed from virus to bacteria.

Properties of Genetic Material:
- It should be able to generate its replica (replication).
- It should chemically and structurally be stable.
- It should provide the scope for slow changes (mutation) that are required for evolution.
- It should be able to express itself in the form of ‘Mendelian Characters’.
- DNA is chemically less reactive but structurally more stable as compare to RNA. So, DNA is better genetic material.
- RNA used as genetic material as well as catalyst and more reactive so less stable. Therefore, DNA has evolved from RNA.
DNA Replication:
The process of producing two identical copies of DNA from a single DNA molecule is called DNA replication. This section describes this whole process in detail, as it is a process of biological inheritance. Students will be familiar with many different terminologies which are associated with the process of DNA replication. You will know about semi-conservative replication, replication fork. This section further explains about leading strand and lagging strand, mentioning the origin of replication at the end of this section.

Transcription:
The process, transcription, where the formation of RNA takes place before the occurrence of gene expression and protein synthesis. You will get an idea about what happens during the transcription process as you read this section. A promoter, structural gene and a terminator are all three different regions in a transcription unit,

Types of RNA and the process of Transcription:
In bacteria, there are three major types of RNAs: mRNA (messenger RNA), tRNA (transfer RNA), and rRNA (ribosomal RNA). All three RNAs are needed to synthesize a protein in a cell. The mRNA provides the template, tRNA brings amino acids and reads the genetic code, and rRNAs play structural and catalytic role during translation.
Transcription in prokaryotes:
Initiation: In this step, RNA polymerase enzyme and initiation factor binds at the promoter of DNA sequence and begin transcription.
Elongation: In this step RNA polymerase, enzyme nucleoside triphosphate behaves as a substrate and polymerases the nucleotides of templates as a complementary strand.
Termination: In this step rho, a terminator factor replaces the initiation factor at the DNA sequence termination point. At this stage RNA and RNA polymerase, an enzyme is separated with rho factor.

Process of Transcription in Eukaryotes:
Three RNA polymerases in the nucleus (in addition to the RNA polymerase found in the organelles). The RNA polymerase I transcribes rRNAs (28S, 18S, and 5.8S), whereas the RNA polymerase III is responsible for transcription of tRNA, 5srRNA, and snRNAs (small nuclear RNAs). The RNA polymerase II transcribes precursor of mRNA, the heterogeneous nuclear RNA (hnRNA). the primary transcripts contain both the exons and the introns and are non-functional. hnRNA undergo two additional processing called as capping and tailing.
Splicing: where the introns are removed, and exons are joined in a defined order.
Capping: In capping an unusual nucleotide (methyl guanosine triphosphate) is added to the 5′-end of hnRNA.
Tailing: In tailing, adenylate residues (200-300) are added at 3′-end in a template independent manner. It is the fully processed hnRNA, now called mRNA, that is transported out of the nucleus for translation.

Exons: Exons are said to be those sequence that appear in mature or processed RNA.
Introns: Introns or intervening sequences do not appear in mature or processed RNA.
The Machinery and the Enzymes:
In living cells, such as E. coli, the process of replication requires a set of catalysts (enzymes). The main enzyme is referred to as DNA-dependent DNA polymerase These enzymes are highly efficient enzymes as they have to catalyse polymerization E. coli that has only 4.6 ×106 bp (compare it with human whose diploid content is 6.6 × 109 bp) Any mistake during replication would result into mutations. energetically replication is a very expensive process.
Replicating Fork:
For long DNA molecules, since the two strands of DNA cannot be separated in its entire length (due to very high energy requirement), the replication occur within a small opening of the DNA helix, referred to as replication fork. The DNA-dependent DNA polymerases catalyse polymerisation only in one direction, that is 5′ → 3′. on one strand (the template with polarity 3′ → 5′), the replication is continuous, while on the other (the template with polarity 5′ → 3′), it is discontinuous. The discontinuously synthesized fragments are later joined by the enzyme DNA ligase The DNA polymerases on their own cannot initiate the process of replication. Also the replication does not initiate randomly at any place in DNA. There is a definite region in E. coli DNA where the replication originates. Such regions are termed as origin of replication. It is because of the requirement of the origin of replication that a piece of DNA if needed to be propagated during recombinant DNA procedures, requires a vector. The vectors provide the origin of replication.

Genetic Code:
- Genetic Code is the relationship of amino acids sequence in a polypeptide and nucleotide/ base sequence in mRNA. It directs the sequence of amino acids during synthesis of proteins.
- George Gamow suggested that genetic code should be combination of 3 nucleotides to code 20 amino acids.
- H.G. Khorana developed chemical method to synthesizing RNA molecules with defined combination of bases.
- Marshall Nirenberg’s cell free system for protein synthesis finally helped the code to be deciphered.

Salient features of Genetic Code are:
- The code is triplet. 61 codons code for amino acids and 3 codons do not code for any amino acids called stop codon (UAG, UGA and UAA).
- Codon is unambiguous and specific, code for one amino acid.
- The code is degenerate. Some amino acids are coded by more than one codon.
- The codon is read in mRNA in a contiguous fashion without any punctuation.
- The codon is nearly universal. AUG has dual functions. It codes for methionine and also act as initiator codon.
Mutations and Genetic code:
A change of single base pair (point mutation) in the 6th position of Beta globin chain of Hemoglobin results due to the change of amino acid residue glutamate to valine. These results into diseased condition called sickle cell anemia. Insertion and deletion of three or its multiple bases insert or delete one or multiple codons hence one or more amino acids and reading frame remain unaltered from that point onwards. Such mutations are called frame-shift insertion or deletion mutations.
tRNA– the Adapter Molecule:
The t-RNA called as adaptor molecules. It has an anticodon loop that has bases complementary to code present on mRNA and also has an amino acid acceptor to which amino acid binds. t-RNA is specific for each amino acids.
The secondary structure of t-RNA is depicted as clover-leaf. In actual structure, the t-RNA is a compact molecule which look like inverted L.

Translation process:
Translation is the process of polymerization of amino acids to form a polypeptide. The order and sequence of amino acids are defined by the sequence of bases in the mRNA. Amino acids are joined by peptide bonds.
It involved following steps:
Charging of t-RNA.
Formation of peptide bonds between two charged tRNA.
The start codon is AUG. An mRNA has some additional sequence that are not translated called untranslated region (UTR).
For initiation ribosome binds to mRNA at the start codon. Ribosomes moves from codon to codon along mRNA for elongation of protein chain. At the end release factors binds to the stop codon, terminating the translation and release of polypeptide form ribosome.

Regulation of Gene Expression:
All the genes are not needed constantly. The genes needed only sometimes are called regulatory genes and are made to function only when required and remain non-functional at other times. Such regulated genes, therefore required to be switched ‘on’ or ‘off’ when a particular function is to begin or stop.
The Lac Operon:
Lac operon consists of one regulatory gene (i) and three structural genes (y, z and a). Gene i code for the repressor of the lac operon. The z gene code for beta-galactosidase, that is responsible for hydrolysis of disaccharide, lactose into monomeric units, galactose and glucose. Gene y code for permease, which increases permeability of the cell. Gene a encode for transacetylase.
Lactose is the substrate for enzyme beta-galactosidase and it regulates switching on and off of the operon, so it is called inducer.
Regulation of Lac operon by repressor is referred as negative regulation. Operation of Lac operon is also under the control of positive regulation.

Human Genome Project (HGP):
Human Genome Project was launched in 1990 to find out the complete DNA sequence of human genome using genetic engineering technique and bioinformatics to isolate and clone the DNA segment for determining DNA sequence.
The project was coordinated by the US Department of Energy and the National Institute of health.
The method involved the two major approaches- first identifying all the genes that express as RNA called Express sequence tags (EST).The second is the sequencing the all set of genome that contained the all the coding and non-coding sequence called sequence Annotation.
Goal of HGP:
- Identify all the genes (20,000 to 25,000) in human DNA.
- Determine the sequence of the 3 billion chemical base pairs that make up human DNA.
- Store this information in data base.
- Improve tools for data analysis.
- Transfer related information to other sectors.
- To address the legal, ethical and social issues that may arise due to project.
Salient features of Human Genome:
- The human genome contains 3164.7 million nucleotide bases.
- The average gene consists of 3000 bases, but sizes vary greatly, with the largest known human gene being dystrophin at 2.4 million bases.
- Less than 2 per cent of the genome codes for proteins.
- Repeated sequences make up very large portion of the human genome.
- Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times.
- Chromosome 1 has most genes (2968), and the Y has the fewest (231).
- Scientists have identified about 1.4 million locations where single base DNA differences (SNPs – single nucleotide polymorphism) occur in humans.
DNA Fingerprinting:
Satellite DNA regions are stretches of repetitive DNA which do not code for any specific protein. These non-coding sequences form a major chunk of the DNA profile of humans. They depict a high level of polymorphism and are the basis of DNA fingerprinting. These genes show a high level of polymorphism in all kind of tissues as a result of which they prove to be very useful in forensic studies.
Any piece of DNA sample found at a crime scene can be analyzed for the level of polymorphism in the non-coding repetitive sequences. After the DNA profile is traced, it becomes easier to find the criminal by performing the DNA fingerprinting for the suspects.
Apart from crime scenes, Fingerprinting applications also prove useful in finding the parents of an unclaimed baby by conducting a paternity test on a DNA sample from the baby.
DNA Fingerprinting Steps:
Alec Jeffreys developed this technique in which he used satellite DNAs also called VNTRs (Variable Number of Tandem Repeats) as a probe because it showed the high level of polymorphism.
Following are the steps involved in DNA fingerprinting:

DNA Fingerprinting Applications:
As discussed earlier the technique of fingerprinting is used for DNA analysis in forensic tests and paternity tests. Apart from these two fields, it is also used in determining the frequency of a particular gene in a population which gives rise to diversity. In case of the change in gene frequency or genetic drift, Fingerprinting can be used to trace the role of this change in evolution.



Class 12 Biology All Chapter Notes
- Chapter 1 Reproduction in Organisms Notes
- Chapter 1 Reproduction in Organisms Question Answer
- Chapter 2 Sexual Reproduction in Flowering Plants Notes
- Chapter 2 Sexual Reproduction in Flowering Plants Question Answer
- Chapter 3 Human Reproduction Notes
- Chapter 3 Human Reproduction Notes Question Answer
- Chapter 4 Reproductive Health Notes
- Chapter 4 Reproductive Health Question Answer
- Chapter 5 Principles of Inheritance and Variation Notes
- Chapter 5 Principles of Inheritance and Variation Question Answer
- Chapter 6 Molecular Basis of Inheritance Notes
- Chapter 6 Molecular Basis of Inheritance Question Answer
- Chapter 7 Evolution Notes
- Chapter 7 Evolution Question Answer
- Chapter 8 Human Health and Disease Notes
- Chapter 8 Human Health and Disease Question Answer
- Chapter 9 Strategies For Enhancement in Food Production Notes
- Chapter 9 Strategies For Enhancement in Food Production Question Answer
- Chapter 10 Microbes In Human Welfare Notes
- Chapter 10 Microbes In Human Welfare Question Answer
- Chapter 11 Biotechnology Principal and Processes Notes
- Chapter 11 Biotechnology Principal and Processes Question Answer
- Chapter 12 Biotechnology and its Applications Notes
- Chapter 12 Biotechnology and its Applications Question Answer
- Chapter 13 Organisms and Populations Notes
- Chapter 13 Organisms and Populations Question Answer
- Chapter 14 Ecosystem Notes
- Chapter 14 Ecosystem Question Answer
- Chapter 15 Biodiversity And Conservation Notes
- Chapter 15 Biodiversity And Conservation Question Answer
- Chapter 16 Environmental Issues Notes
- Chapter 16 Environmental Issues Question Answer